博文
tophat的Max multihits参数设置对gene expression估计的影响
||||

Plots to assess the effect of the parameter 'Max multihits' (X-axis).
'Max multihits' is a TopHat parameter that indicates how many times one tag is permitted to be aligned. (a) Number of genes found by RNAseq (Y-axis) against 'Max multihits'. (b) Spearman's correlation between microarrays (GSM23372, GSM161670, and GSM246123) and RNAseq. (c) Y-axis indicates how many splicing sites found in TopHat were included in known genes or FPKM > 0 genes. (d) Histogram showing the distribution of FPKM. As the number of permitted 'Max multihits' increased, the quantity of genes with small FPKM values increased (black arrows).
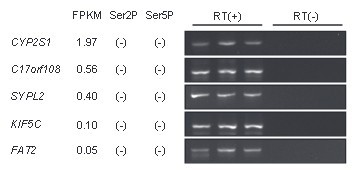
The accuracy of this optimization was reinforced by performing RT-PCR on genes with a small FPKM value (0.05-1.97) when 'Max multihits' was set to 10, and confirming the mRNA expression
结论:
The optimum setting for 'Max multihits' in tophat was 10. This identified the maximum number of genes, saturated the correlation between RNAseq and past expression microarray data sets, and reduced the possibility false positives.
Odawara et al.BMC Genomics 2011 12:516 doi:10.1186/1471-2164-12-516
https://m.sciencenet.cn/blog-981687-733558.html
上一篇:RPS-Blast 安装和使用
下一篇:windows下ssh登录linux图形界面