博文
GWAS综述1(不同血缘群体)
|||
Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations
全基因组关联研究(GWASs)主要关注欧洲血统的人群,但重要的是要更好地代表多样化的人群。增加研究参与者的多样性将促进我们对所有人群遗传结构的理解,并确保遗传研究具有广泛的适用性。为了促进和促进多血统和混合群体的研究,我们概述了主要的方法学考虑,并强调了机会、挑战、解决方案和需要发展的领域。尽管人们认为分析来自不同人群的遗传数据是困难的,但在科学和伦理上这是必须的,而且有一个不断扩大的分 析工具箱来做好它。
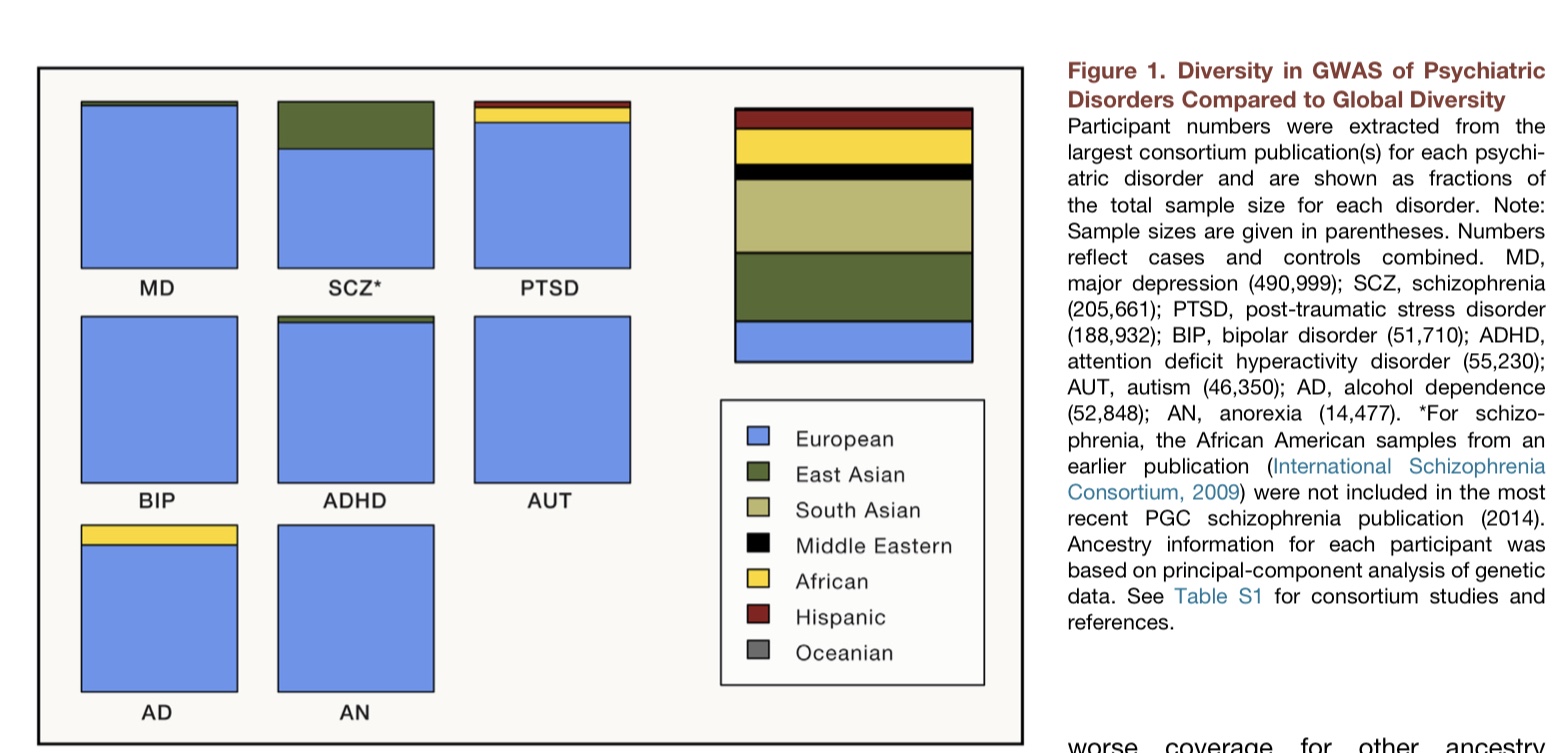
精神疾患的GWAS多样性与全球多样性相比较。主要的项目MD, major depression (490,999); SCZ, schizophrenia (205,661); PTSD, post-traumatic stress disorder (188,932); BIP, bipolar disorder (51,710); ADHD, attention deficit hyperactivity disorder (55,230); AUT, autism (46,350); AD, alcohol dependence (52,848); AN, anorexia (14,477)(图1)。
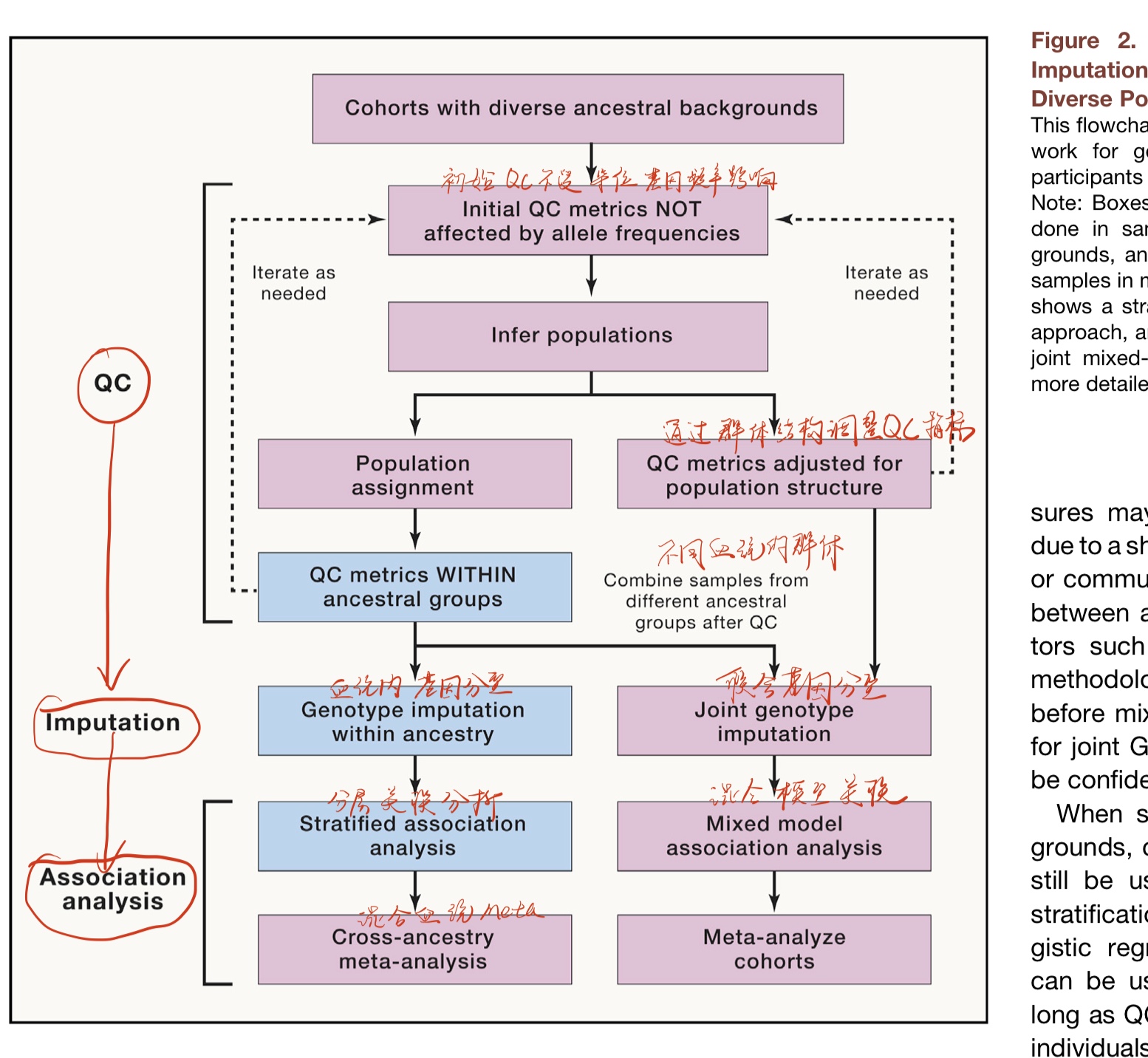
常用的多血统GWAS流程(图2),红色是多血统背景分析流程,蓝色是主要人群组分析流程(单一血统)。左边的是成层(stratified meta-analysis)meta分析策略,右边是联合混合模型策略(joint mixed-model approach)。
本文还综述了目前的方法,陷阱等(table2)。


关于QC的更多细节和方法在附加表中(要钱没看到,希望好心人能分享一下,谢谢)
专用术语
Admixed Population: A population of individuals with ancestors from two or more populations. Admixed can also be used to refer to individuals.
Fine-mapping: Analytical procedures designed to refine GWAS loci to a smaller set of likely causal-variant candidates to facilitate interpretation and follow-up studies.
Genetic Correlation: The correlation of genome-wide genetic effects between two phenotypes, which is often estimated for a subset of genomic variants (e.g. SNPs in a GWAS).
Genotype Imputation: Estimation of genotypes at genetic sites that have not been directly measured, using data from a reference panel to infer genotypes based on LD and haplotype structures. Accuracy depends on availability of suitable reference panels. GWAS: Genome-wide association study. Analysis of common genetic variants across the whole genome for association with a phenotype.
GxE: Gene-by-environment interaction refers to genetic effects on a phenotype that vary based on environment or vice versa. Haplotype: A group of alleles that are correlated with one another because they are inherited together on a chromosome. HWE: Hardy-Weinberg equilibrium, the expected balance of genotypes within a population assuming random mating; infinite population size; and no mutation, migration, or selection. Tests of deviations from HWE are used in quality control to detect technical issues with genotyping. Note that there are also non-technical reasons for deviation from HWE (e.g., selection, population structure, admixture, non-random mating).
LD: Linkage disequilibrium. Alleles in LD are physically linked on a chromosome, which leads to non-random coinheritance such that their frequencies in a population are correlated.
Major Population: A group of individuals with shared genetic ancestry. A heuristic simplification of the complexity of human demography, but useful for describing groups that are likely to have relatively similar allele frequencies and LD patterns due to shared ancestry. Common examples used in practice include continental ancestry groups or ‘‘super populations’’ as defined by the 1000 Genomes Project (e.g., African, Admixed American, East Asian).
PCA: Principal-component analysis. PCA of genotype data is commonly used to examine population structure in a cohort by determining the average genome-wide genetic similarities of individual samples. Derived PCs can be used to group individuals with shared genetic ancestry, to identify outliers, and as covariates, to reduce false positives due to population stratification. Population Stratification: Underlying population structure within a sample that is correlated with a phenotype, which can confound genetic association tests.
PRS: Polygenic risk score. A value computed from an individual’s genotype data that quantifies genetic influences on a particular phenotype; also known as polygenic score (PGS), genetic risk score (GRS), or risk profile score (RPS).
Reference Panel: A set of genetic variants from a population. Reference panels are used to design arrays, impute genotypes, catalogue genetic variants, and identify regions that are similar and different among populations.
SNP Heritability: Proportion of phenotypic variance that is explained by additive genetic effects of a set of SNPs.
文章链接:https://doi.org/10.1016/j.cell.2019.08.051
https://m.sciencenet.cn/blog-3422766-1222767.html
上一篇:nature:睡莲基因组和被子植物早期演化文章阅读笔记
下一篇:关于已知抗原的结构及表位预测