博文
测序数据质量控制:多样本的fastqc结果,一目了然!
|||
The analysis report of quality control of fastq files
The quality control of fastq data produced by high throughput sequencers is performed by FastQC software. FastQC aims to provide a QC report which can spot problems which originate either in the sequencer or in the starting library material. A series of analysis modules are performed by FastQC as shown in the figure below. A ‘normal’ sample as far as FastQC is concerned is random and diverse. Some experiments may be expected to produce libraries which are biased in particular ways. You should treat the summary evaluations therefore as pointers to where you should concentrate your attention and understand why your library may not look random and diverse. Here are URLs of two example reports for illumina data: the good example report and the bad example report.
Specific guidance on how to interpret the output of each module are summarized below.
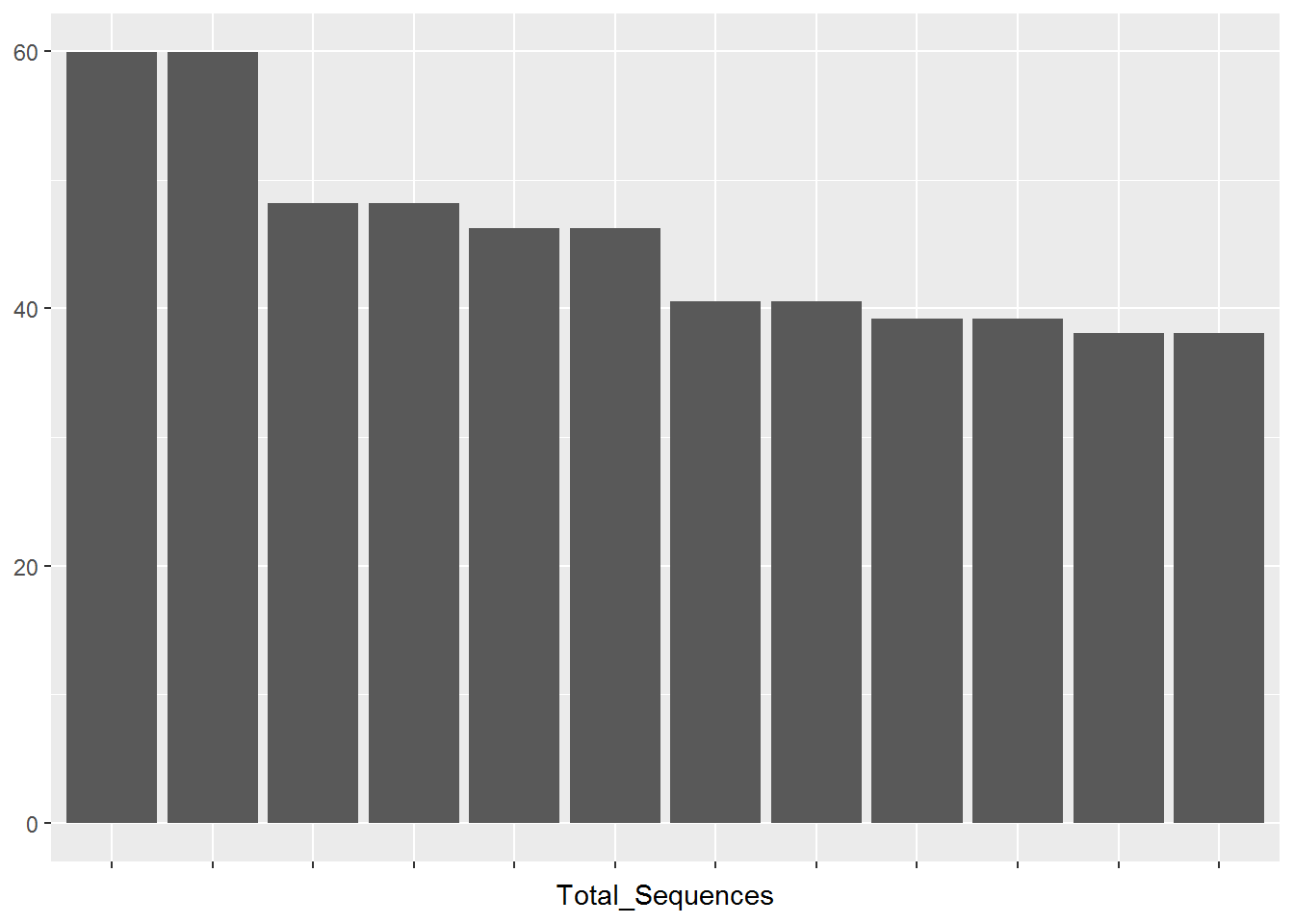
12 fastq files were processed by FASTQC software.
FASTQ files have the same sequence length (150 bases)
FASTQ files have the same quality score Encoding (Sanger / Illumina 1.9)
All the fastq files pass the base sequence quality check
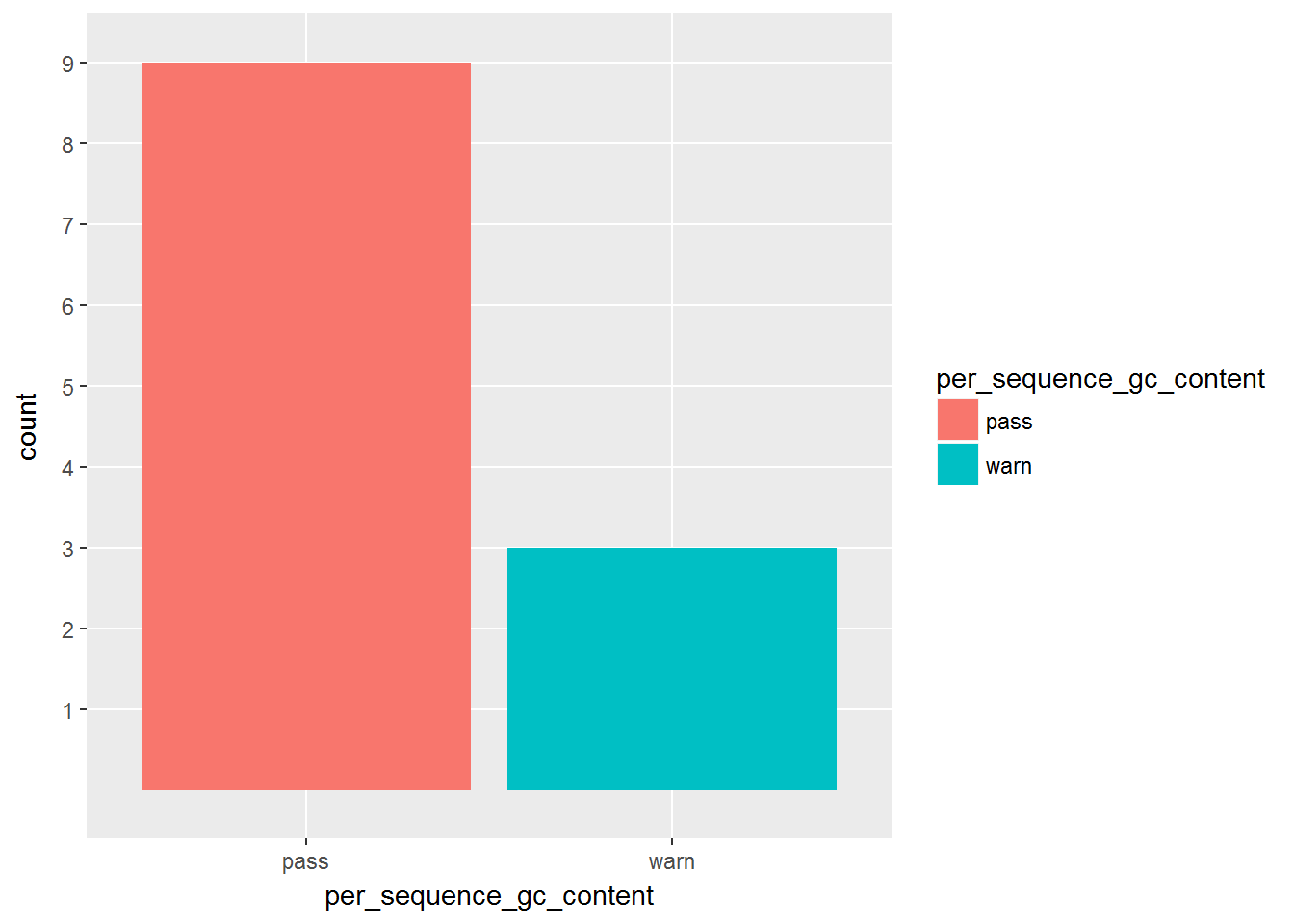
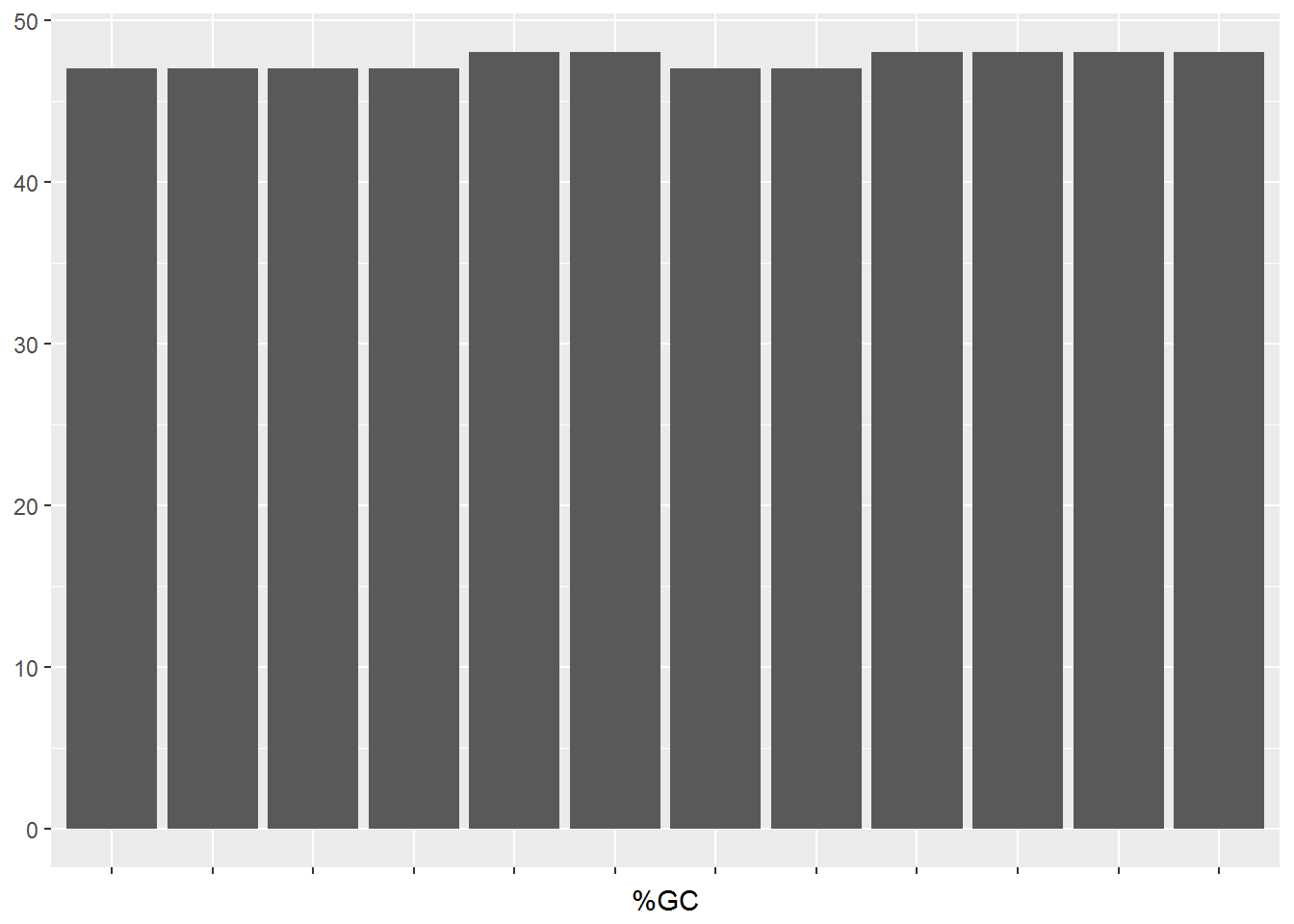
Not all the fastq files pass the gc content check
All the fastq files pass the adapter content check
All the fastq files pass overrepresented sequences check

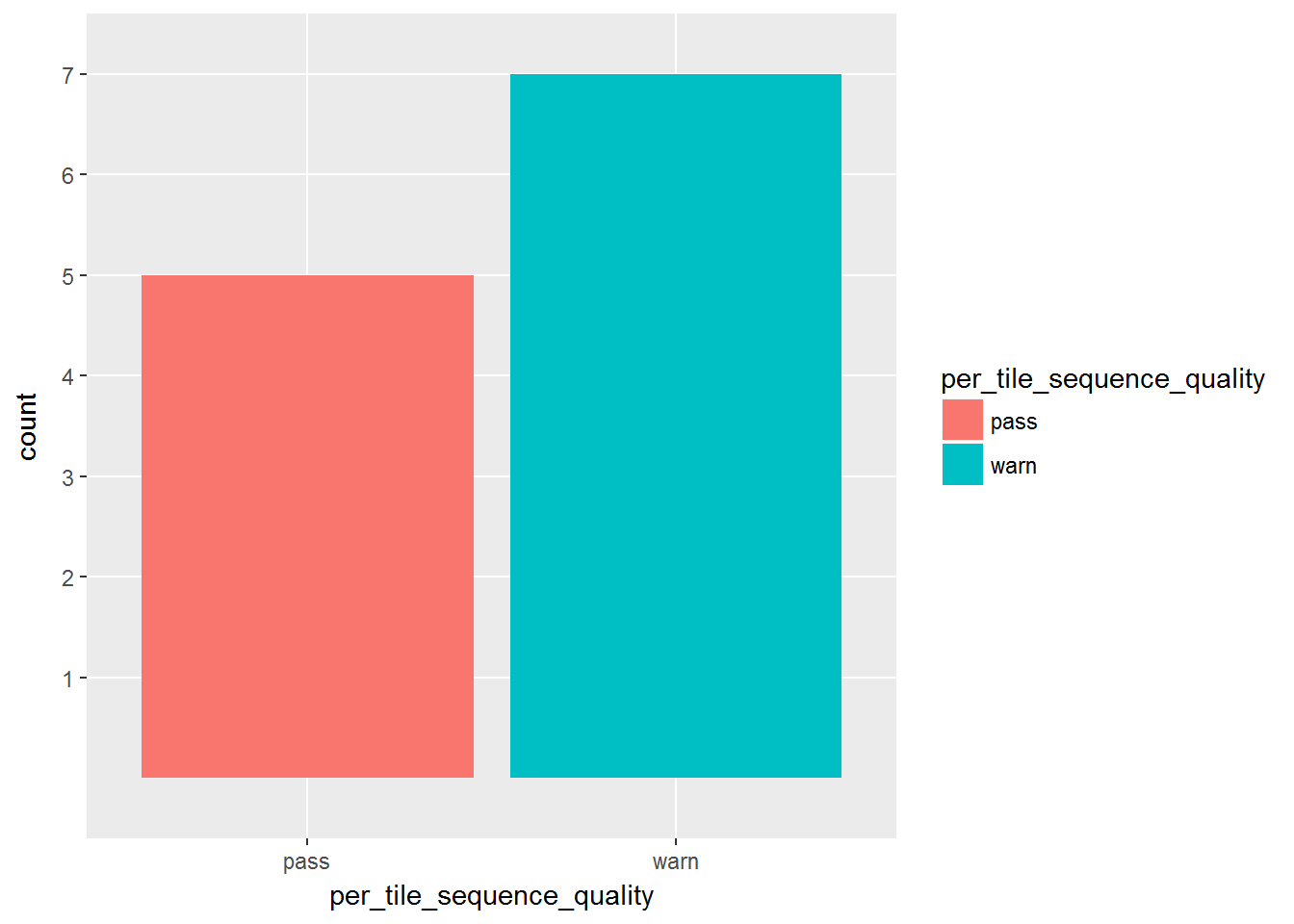
2. Results of analysis modules which are not consistent across all the fastq files

https://m.sciencenet.cn/blog-3291578-1065423.html
上一篇:《实验医学研究导论》阅读笔记
下一篇:用于聚类验证的R包:clValid